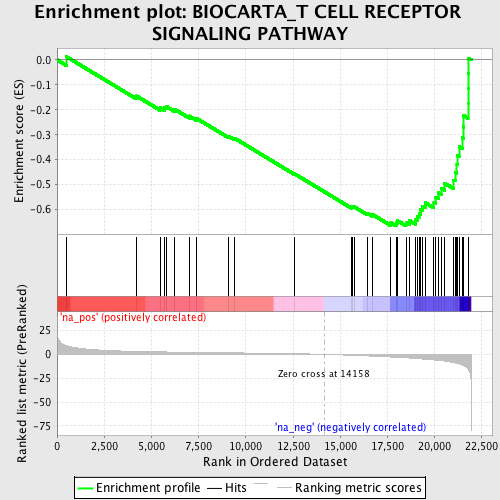
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | BIOCARTA_T CELL RECEPTOR SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.665611 |
| Normalized Enrichment Score (NES) | -2.2253149 |
| Nominal p-value | 0.0 |
| FDR q-value | 8.11735E-5 |
| FWER p-Value | 0.0010 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HRAS | 510 | 8.966 | 0.0120 | No | ||
| 2 | ARHGAP5 | 4207 | 3.039 | -0.1447 | No | ||
| 3 | CAMK2B | 5454 | 2.573 | -0.1915 | No | ||
| 4 | NCF2 | 5673 | 2.503 | -0.1916 | No | ||
| 5 | SOS1 | 5789 | 2.466 | -0.1871 | No | ||
| 6 | PIK3R1 | 6230 | 2.327 | -0.1980 | No | ||
| 7 | RAC1 | 7002 | 2.119 | -0.2249 | No | ||
| 8 | ARHGAP6 | 7368 | 2.020 | -0.2336 | No | ||
| 9 | PPP3CA | 9099 | 1.601 | -0.3063 | No | ||
| 10 | PPP3CC | 9402 | 1.530 | -0.3140 | No | ||
| 11 | JUN | 12565 | 0.666 | -0.4558 | No | ||
| 12 | GRB2 | 15610 | -0.952 | -0.5910 | No | ||
| 13 | LAT | 15646 | -0.971 | -0.5888 | No | ||
| 14 | MAP2K1 | 15741 | -1.040 | -0.5890 | No | ||
| 15 | ZAP70 | 16444 | -1.608 | -0.6147 | No | ||
| 16 | SHC1 | 16723 | -1.857 | -0.6201 | No | ||
| 17 | ELK1 | 17690 | -2.773 | -0.6532 | No | ||
| 18 | MAPK3 | 17962 | -3.067 | -0.6535 | Yes | ||
| 19 | ARFGAP3 | 18028 | -3.129 | -0.6442 | Yes | ||
| 20 | ARHGAP1 | 18490 | -3.648 | -0.6509 | Yes | ||
| 21 | RALBP1 | 18685 | -3.861 | -0.6445 | Yes | ||
| 22 | PRKCA | 18987 | -4.255 | -0.6415 | Yes | ||
| 23 | RAF1 | 19074 | -4.383 | -0.6281 | Yes | ||
| 24 | FOS | 19178 | -4.546 | -0.6149 | Yes | ||
| 25 | ARFGAP1 | 19249 | -4.646 | -0.5998 | Yes | ||
| 26 | CD3G | 19364 | -4.837 | -0.5860 | Yes | ||
| 27 | PTPRC | 19517 | -5.062 | -0.5730 | Yes | ||
| 28 | CD3D | 19969 | -5.819 | -0.5707 | Yes | ||
| 29 | VAV1 | 20066 | -6.018 | -0.5513 | Yes | ||
| 30 | PLCG1 | 20195 | -6.259 | -0.5325 | Yes | ||
| 31 | PPP3CB | 20348 | -6.597 | -0.5135 | Yes | ||
| 32 | PIK3CA | 20532 | -7.036 | -0.4941 | Yes | ||
| 33 | RELA | 21022 | -8.671 | -0.4823 | Yes | ||
| 34 | ARHGAP4 | 21091 | -8.891 | -0.4504 | Yes | ||
| 35 | CD4 | 21188 | -9.300 | -0.4181 | Yes | ||
| 36 | MAPK8 | 21208 | -9.408 | -0.3820 | Yes | ||
| 37 | FYN | 21301 | -9.911 | -0.3471 | Yes | ||
| 38 | MAP2K4 | 21466 | -10.991 | -0.3113 | Yes | ||
| 39 | NFATC1 | 21534 | -11.557 | -0.2689 | Yes | ||
| 40 | CD3E | 21545 | -11.647 | -0.2234 | Yes | ||
| 41 | NFKBIA | 21775 | -15.114 | -0.1744 | Yes | ||
| 42 | LCK | 21778 | -15.166 | -0.1147 | Yes | ||
| 43 | CD247 | 21783 | -15.293 | -0.0547 | Yes | ||
| 44 | MAP3K1 | 21803 | -15.769 | 0.0066 | Yes |